Software
For my research and duties at the Spinoza Centre for Neuroimaging (location Roeterseiland), I have developed several software packages (of which most using Python). You can find a selection below, but see my personal Github and the Spinoza Centre’s Github for a complete overview.
Medusa (work-in-progress!)
Medusa is a Python toolbox to perform 4D face reconstruction and analysis. You can use it to reconstruct a series of 3D meshes of (moving) faces from video files: one 3D mesh for each frame of the video (resulting in a “4D” representation of facial movement). In addition to functionality to reconstruct faces, Medusa also contains functionality to preprocess and analyze the resulting 4D reconstructions.
noiseceiling
In machine learning analyses, researchers often try to maximize cross-validated model performance (e.g., R-squared or classification accuracy). Due to measurement noise, however, the theoretical maximum performance score is not the same as the optimal performance score. I wrote some code that estimates such a noise ceiling for any regression model (assuming R-squared is used) or classification model (any metric), available as a Python package here.
bidsify
Widespread sharing of neuroimaging data is, unfortunately, not common in the neuroscience community. In the past years, some researchers have proposed a common format for MRI datasets - the Brain Imaging Data Structure (BIDS) - which aims at improving data sharing, reproducibility, and transparency through providing a guidelines on how to organize your data (which can be, subsequently, be uploaded to openneuro.org very easily).
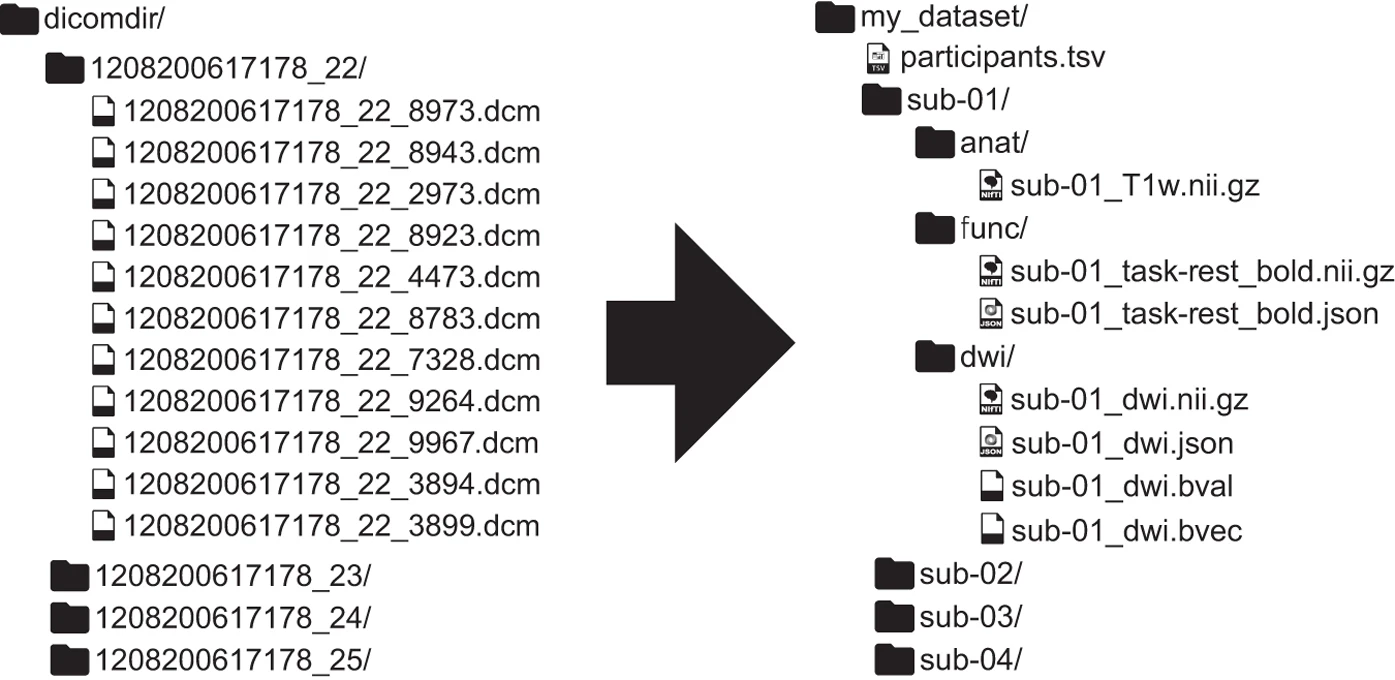 Image from https://www.nature.com/articles/sdata201644
Image from https://www.nature.com/articles/sdata201644
Organizing your raw data into the required BIDS format, however, can be painstakingly time-consuming: you’ve to convert your files in the right format (e.g. Philips PAR/REC or DICOMs to compressed nifti), rename your files, and extract the necessary meta-data. I’ve written a small package that automatically converts your “raw” and unstructured dataset to BIDS, given a config file that you provide (such that the software “knows” which how to convert/format/rename your files). Recently, I also provide a Docker image so that you can run bidsify without worrying about dependencies. The package has been tested on several datasets recorded at the Philips 3T and 7T scanners from the Spinoza Centre for Neuroimaging, and is currenlty deployed at our MRI center to automatically convert all raw scanner data to BIDS (as part of the nitools package, see below).
The code can be found on Github. The README should contain enough information to get started with bidsify.
scanphyslog2bids
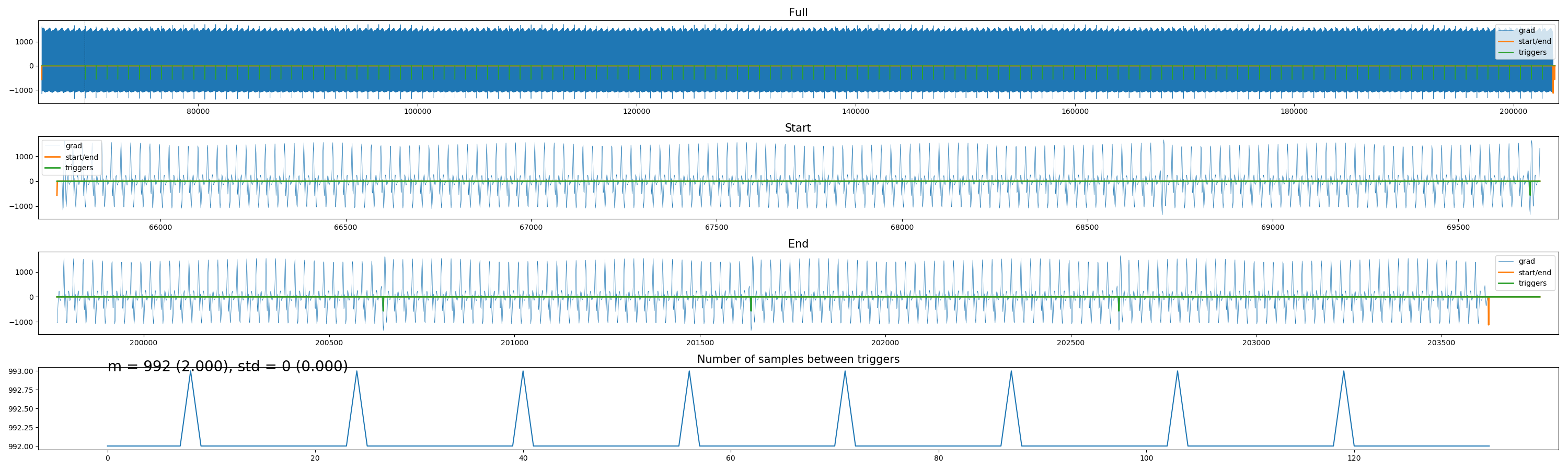
Ever tried to distill volume triggers from Philips physiology (“SCANPHYSLOG”) files? Trust me, it’s frustrating. I created a small package to do this using either trigger markers, or based on the scanner’s gradients (if they’re logged), or, if everything else fails, to interpolate volume triggers “counting backwards” from the offset of the acquisition. Check it out here.
exptools2
Many experimental paradigms in psychology/neuroscience are created programmatically in order to have strict control over the order, timing, and logging of stimuli and responses. Together with people from the lab of Tomas Knapen, I developed the exptools2 package, which is basically a wrapper around the software package PsychoPy. With exptools, we provide a set of routines that ensure precise timing and logging of stimulus onset and responses (as well as Eyetracker integration), which is crucial for many neuroimaging experiments. Check out the repository (and especially the README) to get started!
skbold: Utilities and tools for machine learning on BOLD-fMRI data
Inspired by the scikit-learn (often abbreviated as sklearn) machine learning package in Python, I created skbold - a package aimed at complementing scikit-learn in the organization, representation, and (pre)processing of fMRI data for machine learning (“decoding”) analyses. The package aims for flexibility in the sense that the annoying part of building decoding pipelines (such as getting it in the right format, keeping track of model performance) is taken care of, while the users can still build the actual pipelines using scikit-learn functionality.
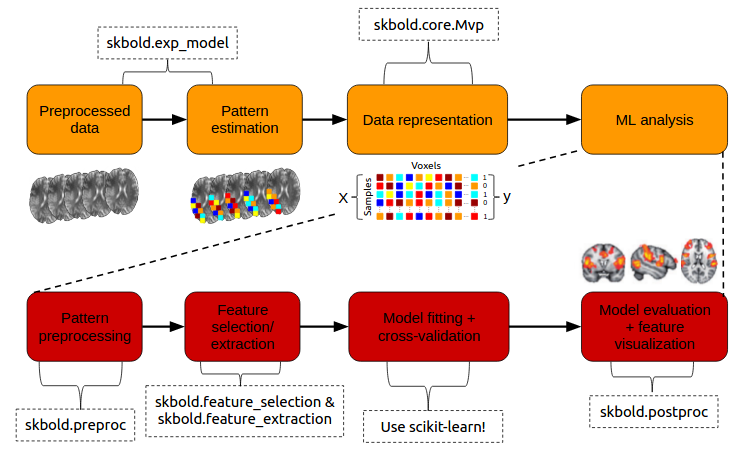
The code can be found on Github and the documentation on ReadTheDocs.
Porcupine
While working on Spynoza during the BrainHack organized by the Spinoza centre, I got to know Tim van Mourik from the Donders Institute, who was working on an open-source pipelining package with a graphical user interface named “Porcupine”, which is short for “PORcupine Creates Ur PipelINE” - in Tim’s words “the worst self-referencing acronym with bad capitalisation and annoying use of slang.” Together with Tomas Knapen, we started working together on Porcupine.
In short, Porcupine is a graphical application that allows you build processing pipelines by defining “nodes” - that implement some function (such as an MRI-processing step) - and connections between them, and importantly then is able to generate the code that would be needed to actually run the pipeline as defined in Porcupine! For example, one framework Porcupine supports is the amazing Nipype framework. So, you could for example build a Porcupine-pipeline implementing an MRI-preprocessing pipeline based on “nodes” from different software packages (such as FSL, AFNI, and Freesurfer) and subsequenly let Porcupine generate the (Nipype-based) code that you can actually run!
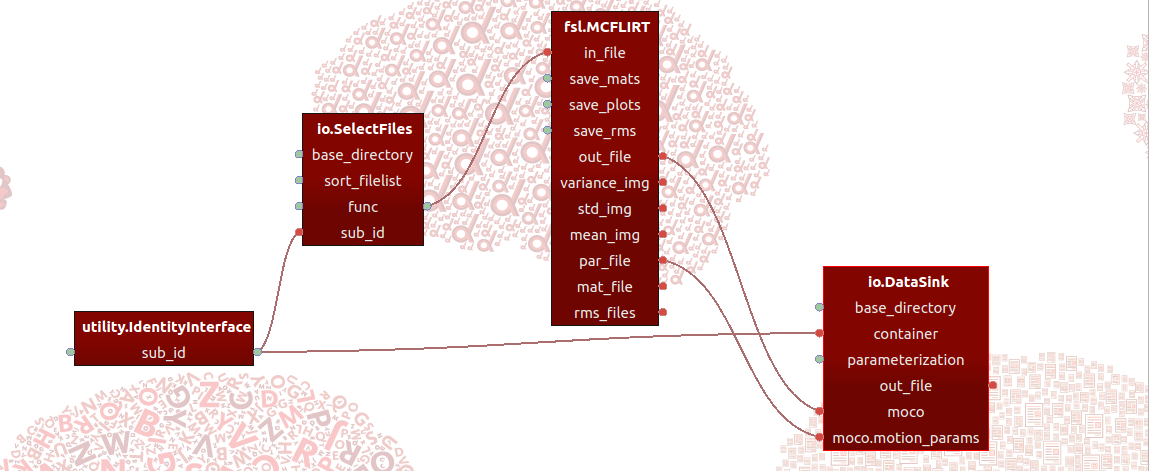
You can find extensive documentation and download links here.
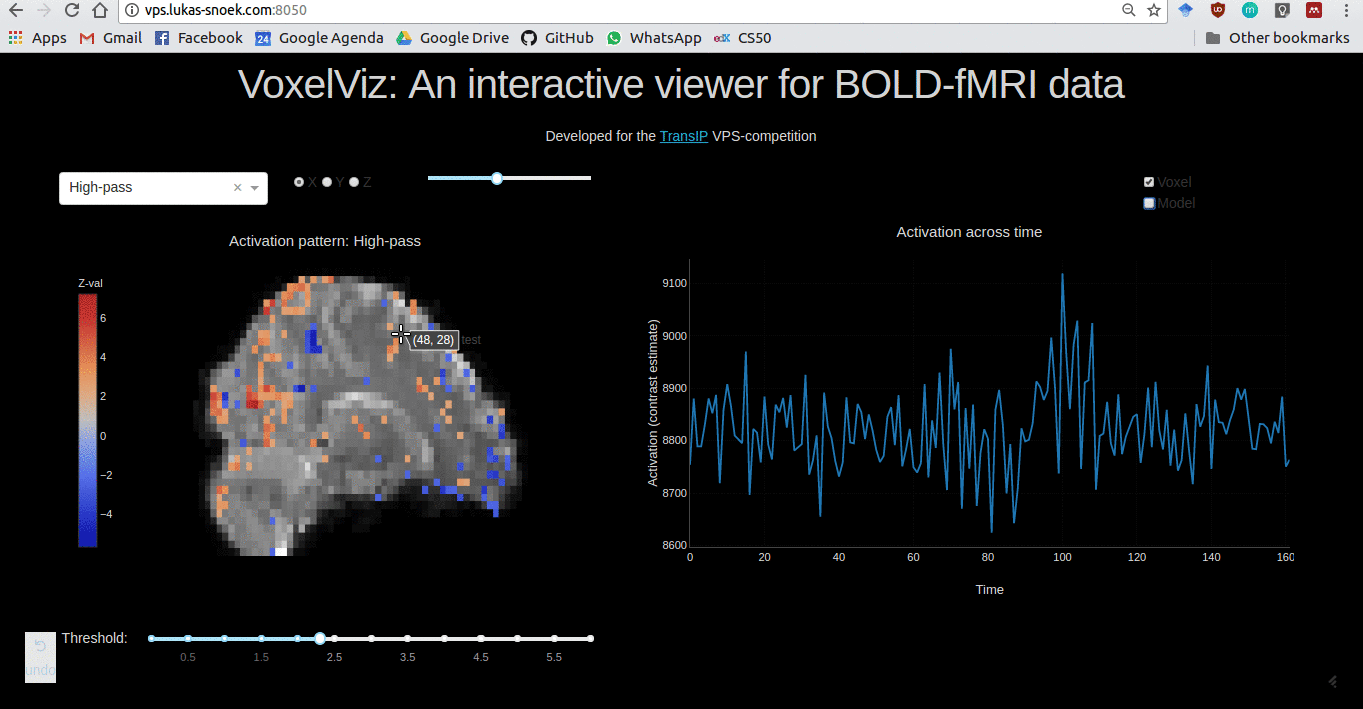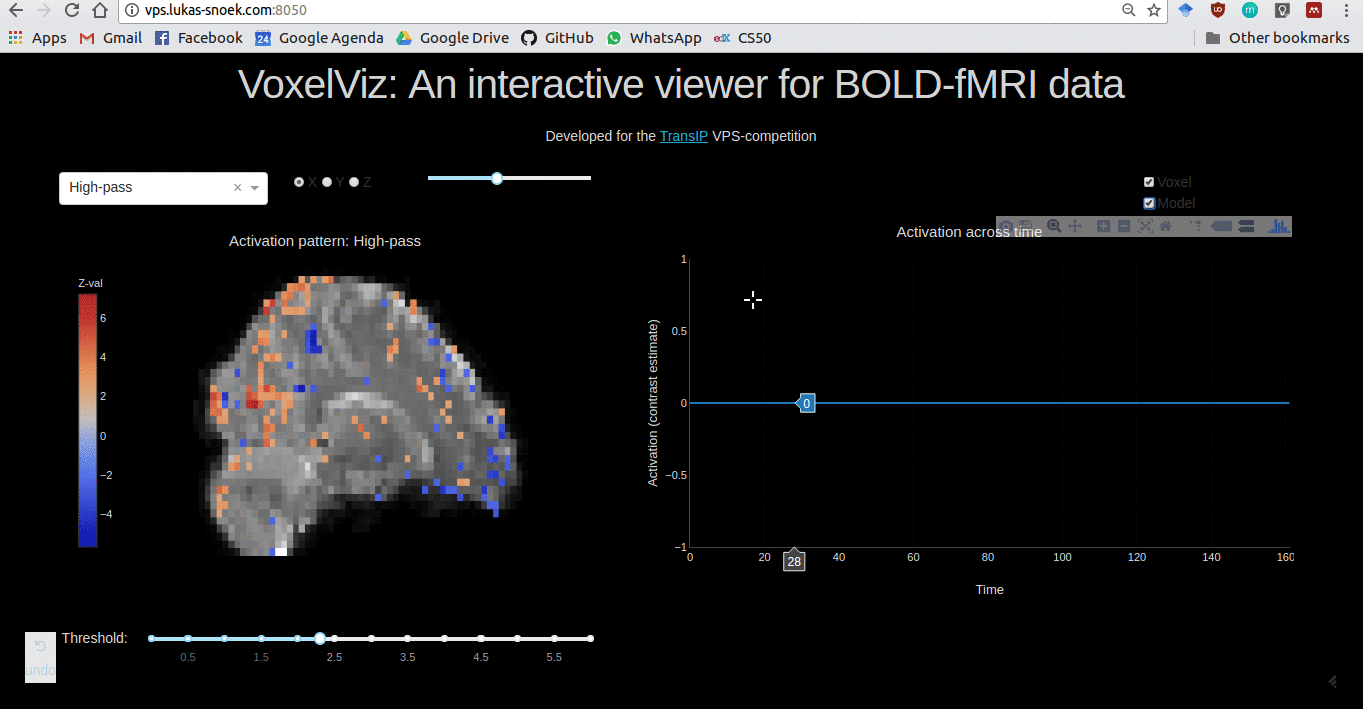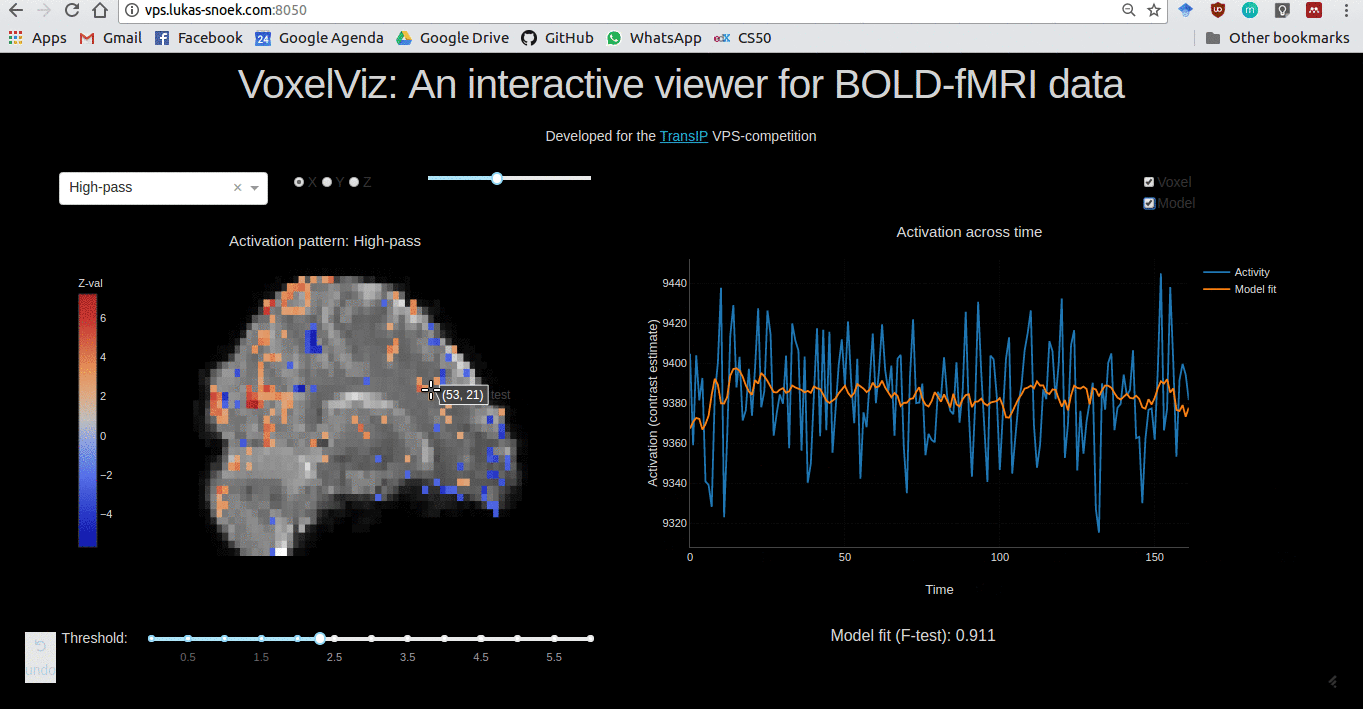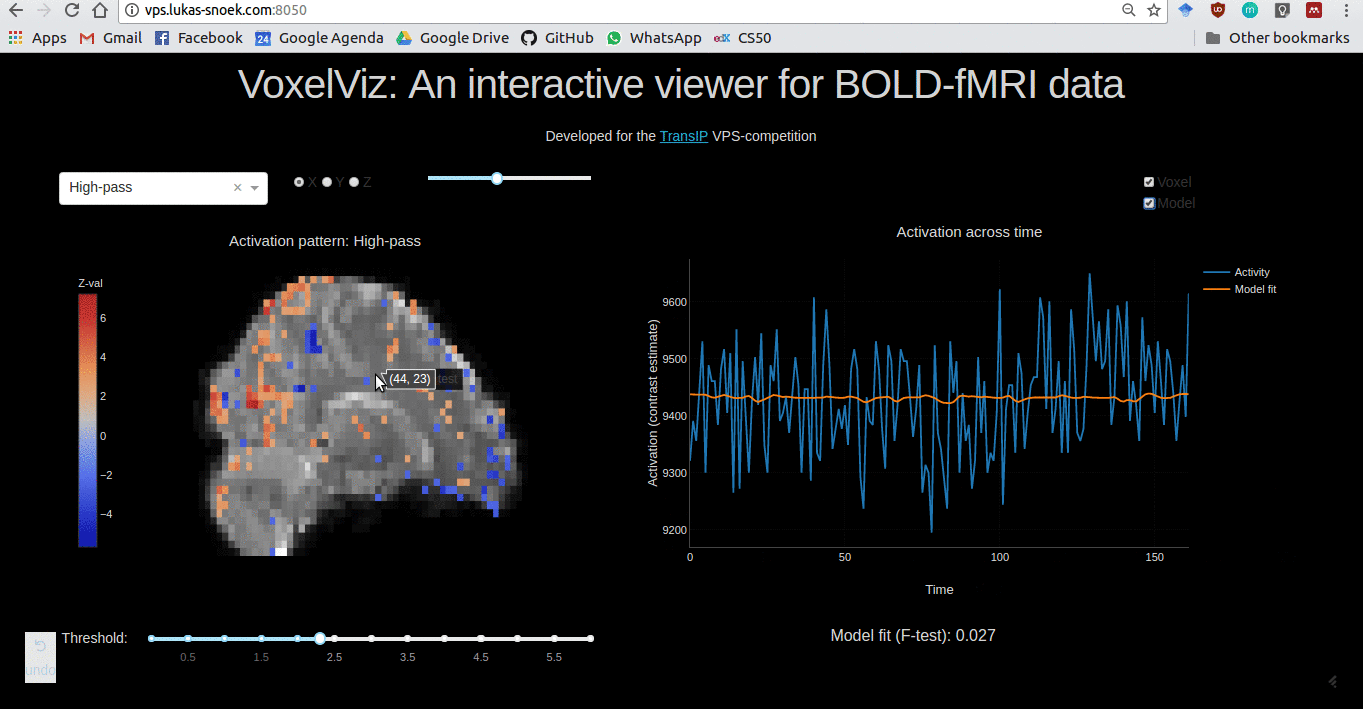
VoxelViz
For the VPS-competition (“Develop an original application for a virtual private server”) by TransIp, I’ve developed an online MRI-viewer — “VoxelViz”. While it’s offline now, the code can be viewed here.